Genome wide Association
Genome wide Association studies for heat stress tolerance
An association mapping panel (HTAM panel) was formed combining the advanced lines from CIMMYT and other contributing partners for the purpose of association mapping to identify genomic regions contributing to heat stress tolerance with the following composition.
| Source | No. Lines |
| CIMMYT-Asia | 343 |
| DTMA | 106 |
| MMRI – Pakistan | 52 |
| Purdue | 23 |
| Kaveri – India | 10 |
| Total | 534 |
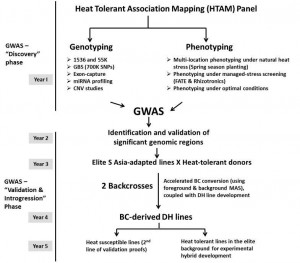
The lines were test crossed to CML451 (HG-B) and CL02450 (HG-A) and TCs phenotyped under heat stress and optimal conditions in >10 locations in 2013 and 2014 summer season. The panel was genotyped in high density using Genotyping by Sequencing (GBS), which yielded ~900K SNPs. The scheme followed for association mapping and further applications is depicted below.
A total of 16 haplotype blocks were identified across testers and locations that were associated with yield under heat stress, whereas 10 haplotype blocks were identified for ASI under heat stress as below. Of these six were tester independent for yield under heat stress and one was found to be tester independent for ASI under heat stress.
| Chr | Position (AGP_V2) | # SNPs |
| GY_Heat | ||
| Tester independent | ||
| 2 | 222.08-222.27 | 7 |
| 7 | 168.29-168.46 | 5 |
| 9 | 98.89-99.08 | 6 |
| 10 | 148.41-148.62 | 5 |
| 3 | 3.43-3.99 | 7 |
| 10 | 148.29-148.62 | 6 |
| Tester1 | ||
| 2 | 44.18-44.38 | 3 |
| 3 | 219.78-220.02 | 5 |
| 4 | 2.60-2.76 | 3 |
| 3 | 3.69-3.99 | 5 |
| 3 | 219.78-220.02 | 5 |
| Tester2 | ||
| 3 | 183.86-184.12 | 5 |
| 7 | 162.09-162.33 | 6 |
| 9 | 98.89-99.08 | 5 |
| 2 | 6.15-6.33 | 3 |
| 9 | 98.89-99.08 | 5 |
| ASI_Heat | ||
| Tester independent | ||
| 1 | 6.77-6.83 | 7 |
| Tester1 | ||
| 1 | 6.77-6.80 | 6 |
| 5 | 190.95-191.08 | 3 |
| Tester2 | ||
| 1 | 187.58-187.66 | 6 |
| 1 | 295.44-295.76 | 3 |
| 2 | 188.85-188.90 | 3 |
| 2 | 202.28-202.42 | 3 |
| 2 | 205.94-206.12 | 4 |
| 2 | 217.65-219.68 | 6 |
| 3 | 199.25-199.27 | 3 |